Microbiome Gym provides 10 statistical tests for the differential abundance analysis of microbiome count data
T-test and ANOVA
Wilcoxon rank-sum test and Kruskal-Wallis test
DESeq2
edgeR
WaVE-DESeq2 and WaVE-edgeR
MetagenomeSeq
DA test
corncob
LDM
ZINB-DPP
We provided a unified interface to use any of the analysis methods. The details can be accessed in github
Extensive analysis can be achieved by utilizing the results curated in MicrobiomeGym. We provide two examples to illustrate use cases.
Example 1: Understanding the performance of an existing MWAS methods.
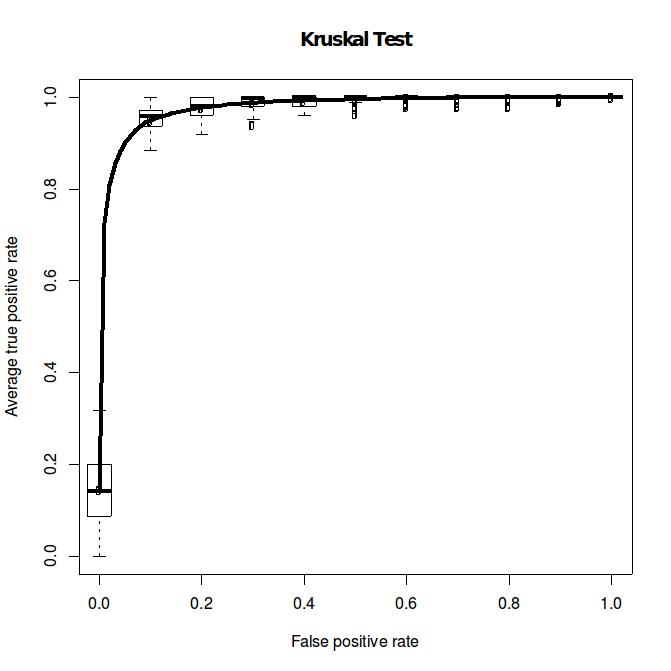
The Wilcoxon ranksum test is arguable the mostly used statistical test used in MWAS. Let’s
examine its overall performance for moderate effect size (effect size = 1) and small sample
size (n = 24). Assume the observe datasets have moderate sparsity, we can report the
performance of Wilcoxon ranksum test using the simulated datasets from Dirichlet multinomial
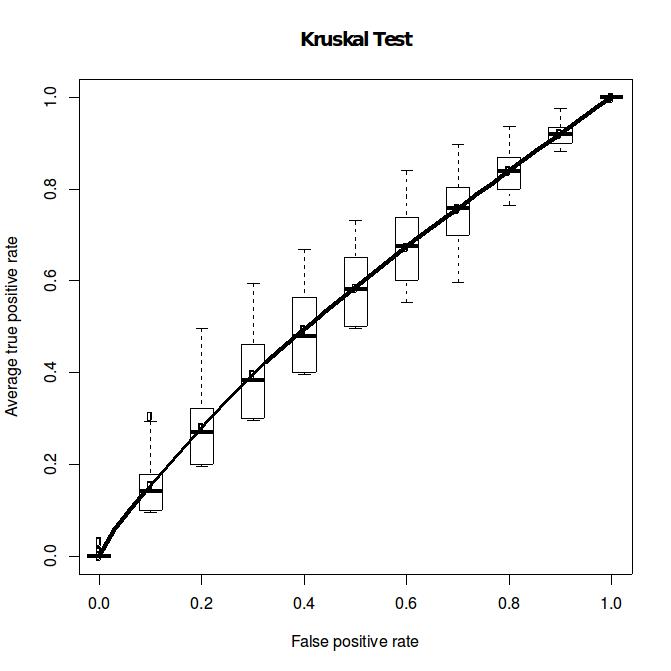
distribution (left, DM_K=2_n=24_1_Kruskal.pdf). However, what is the dataset have large
proportion of zero? The right figure (ZINB_K=2_n=24_1_Kruskal.pdf) shows the perform can
degrade to an impressive extent. It may be good to consider alternative statistics
tests.
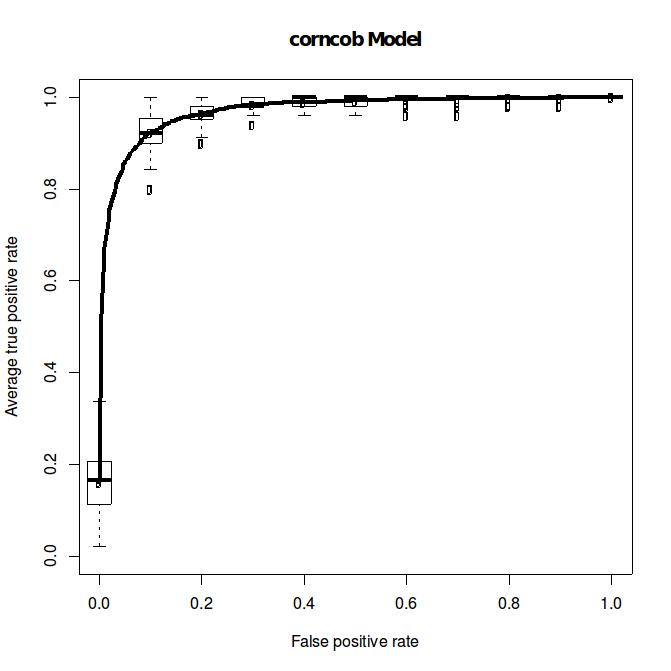
Example 2: A new published method (corncob) is reported with good performance. Will it perform well in a similar scenario to Example 1? A quick way to check is from the ROC plot (DM_K=2_n=24_1_corncob.pdf), which shows improvement over the Wilcoxon ranksum test. Note that we do not require to reproduce the simulation reported in the paper, as MicrobiomeGym already curated simulated datasets to help you evaluate methods. Additionally the codes to reproduce the ROC results (or apply to your own datasets) is available to use too.