Our Tools
HD-STAINING ANALYSIS TOOL
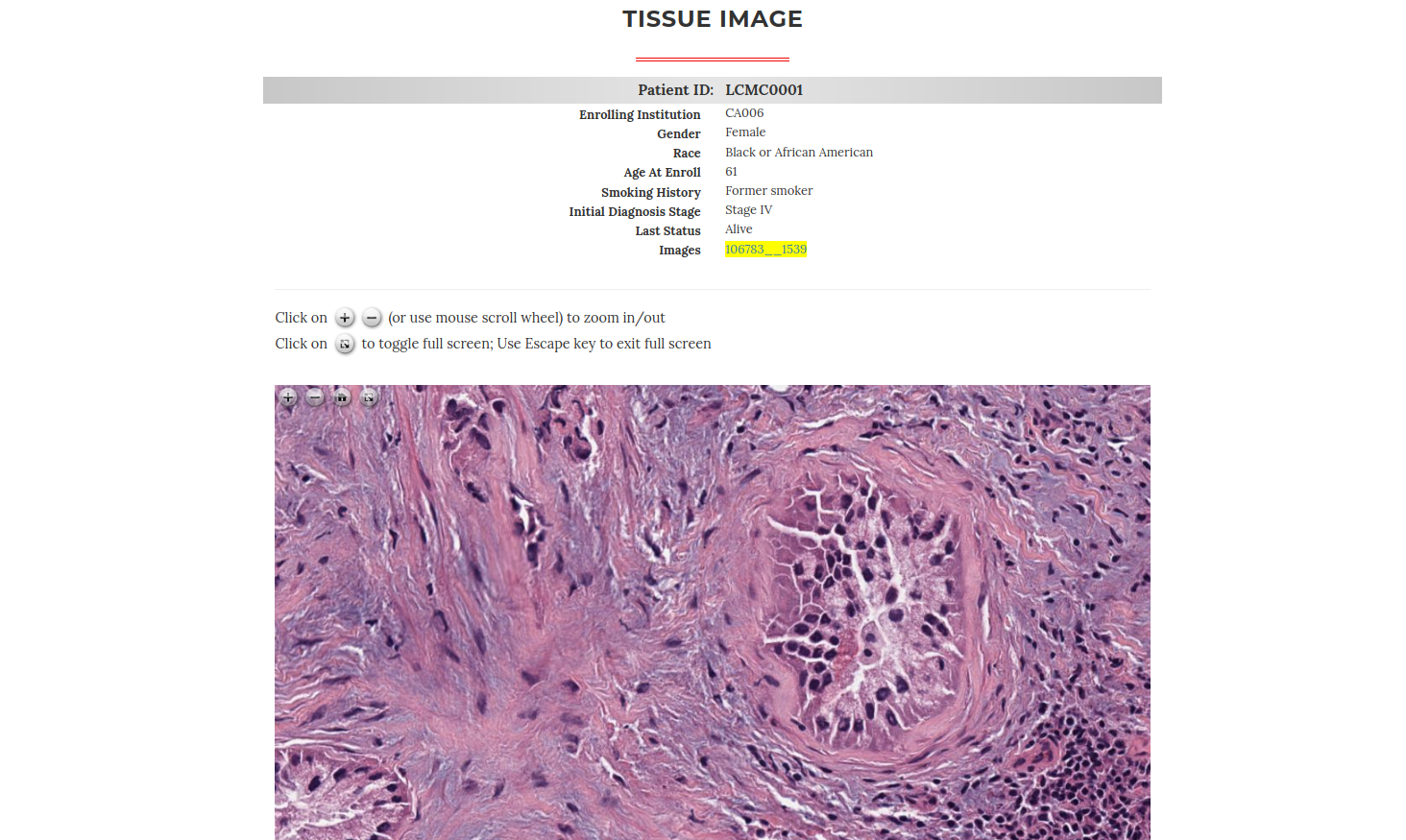
The HD-Staining algorithm is implemented as an online image segmentation tool to provide a user-friendly approach. After simply submitting a H&E stained pathology image, the user will be able to view and download the computational staining results together with nuclei characteristics.
View the websiteHepatic Ploidy Analysis
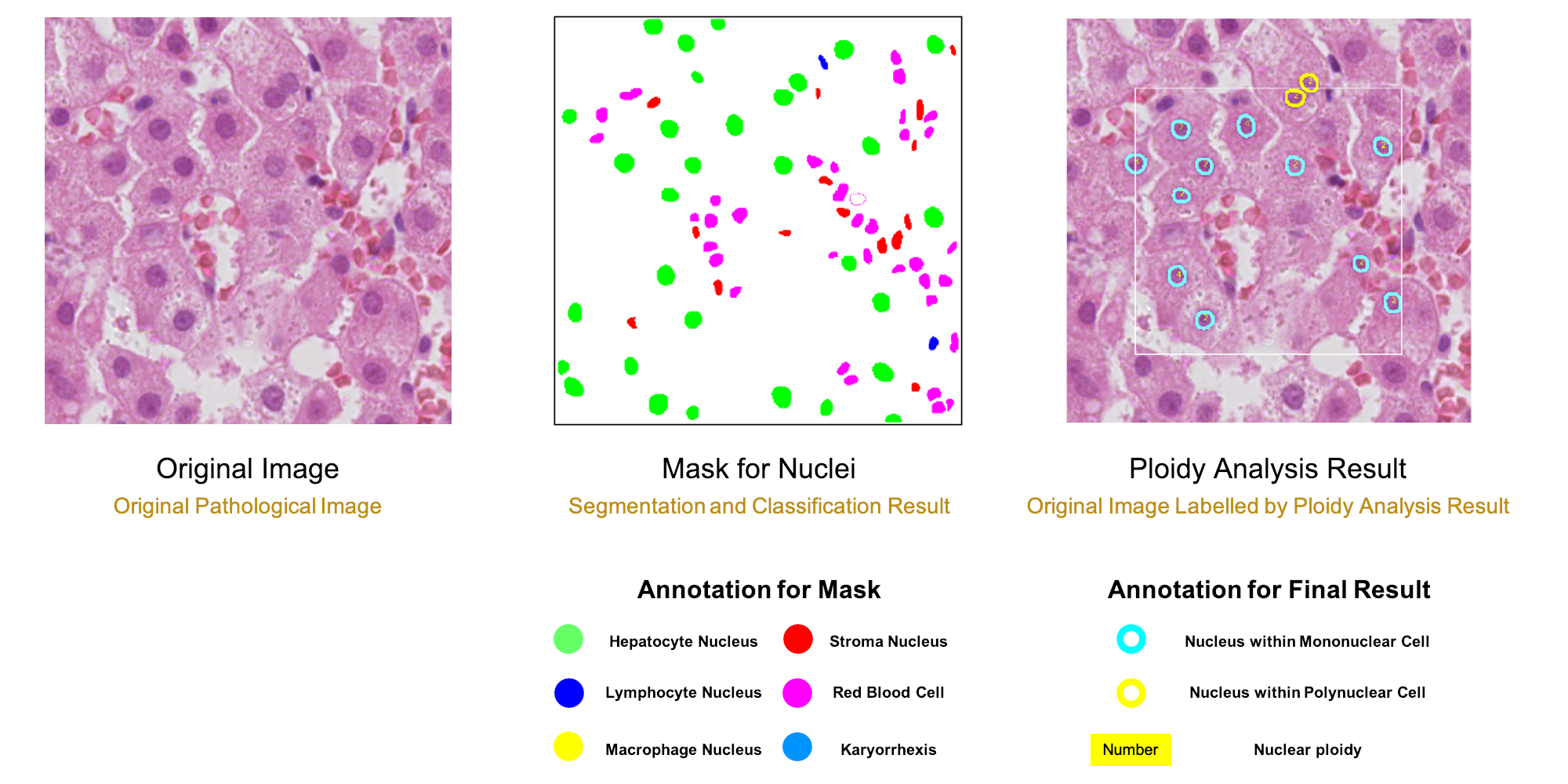
To make the whole analysis procedure more accessible for clinical samples, the algorithm can quantify ploidy information using hematoxylin-eosin (H&E) histopathology slides. A deep-learning model was trained to segment and classify different types of nuclei from H&E histopathology images. Based on the identified hepatocyte nuclei, both cellular and nuclear ploidy are calculated.
View the websiteSingle cell Spatial Profiling Viewer
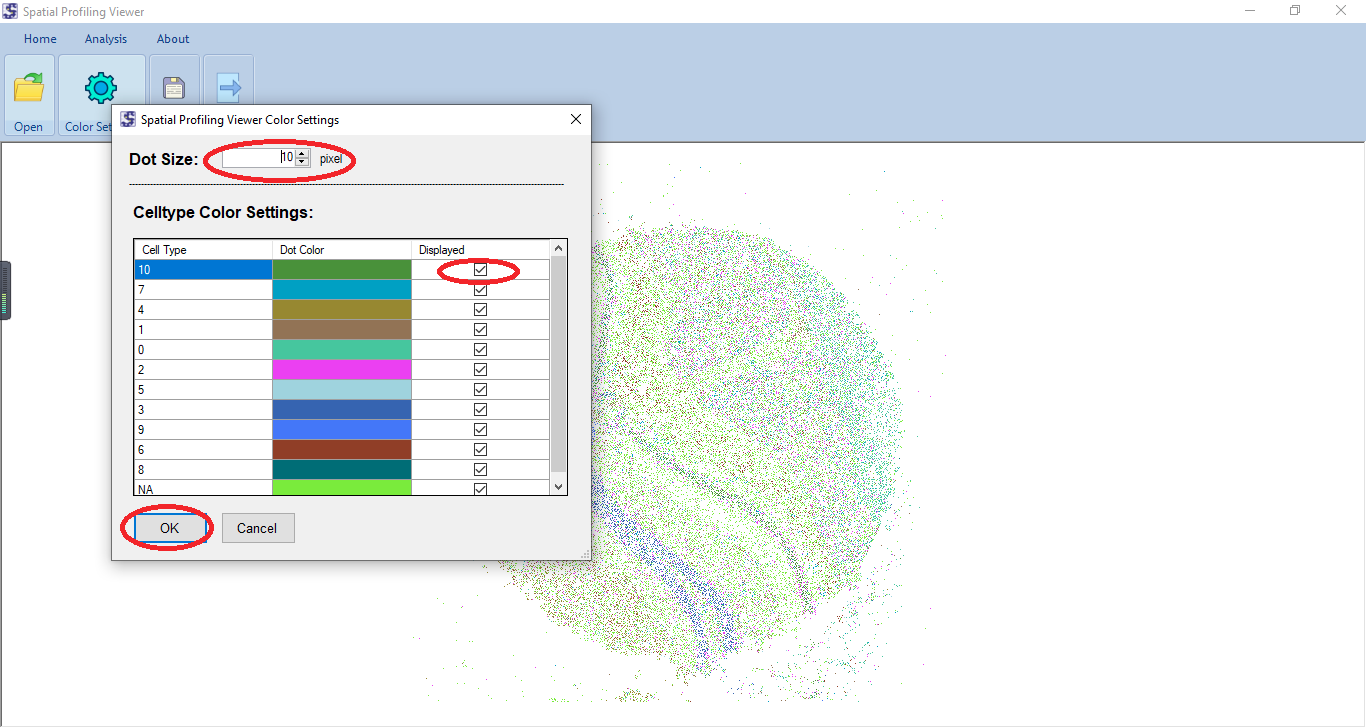
We present a standalone tool, Spatial Profiling Viewer (SPV), to visualize and analyze wide-range of single cell spatial transcriptomics datasets. The software requires plain input data: the cells Cartesian coordinates and cell types. It offers four categories of functions
View the websiteCNN Predicts Prognosis of Lung Adenocarcinoma Patients
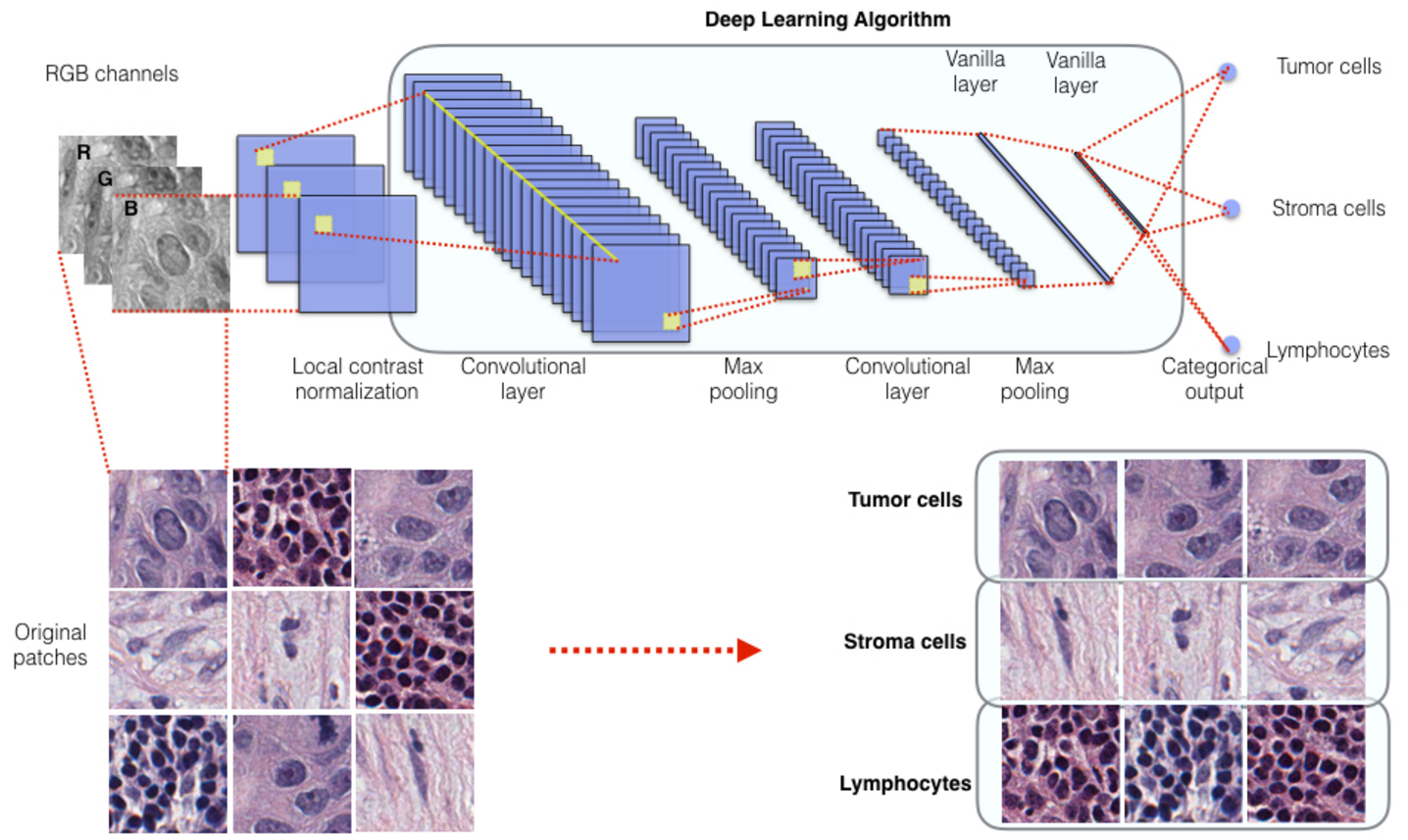
Human evaluation is objective and labor-intensive. In addition, pathological images may contain important prognosis and treatment response information that cannot be reliably understood by humans. Therefore, it is a very important to create machine learning algorithms to automatically recognize pathological imaging data and correlate the automatically retrieved features with patient phenotype.
View the websitePathology Image Visulization and Analysis
We present users a web-based viewer for high-resolution zoomable images. It can be easily integrated into any web applications which provides many viewer functions such as zoom in & zoom out, drag & draw
View the websiteShape Analysis Tool
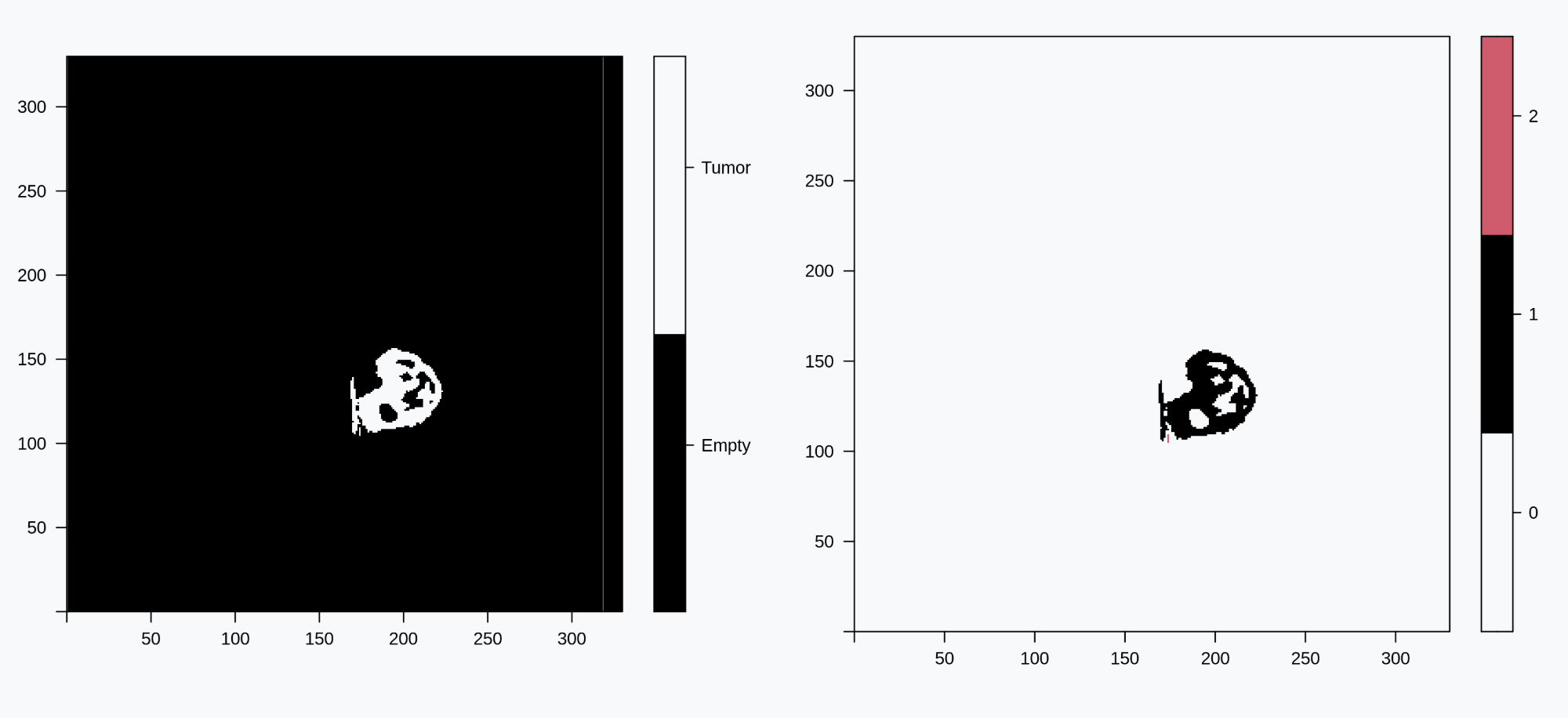
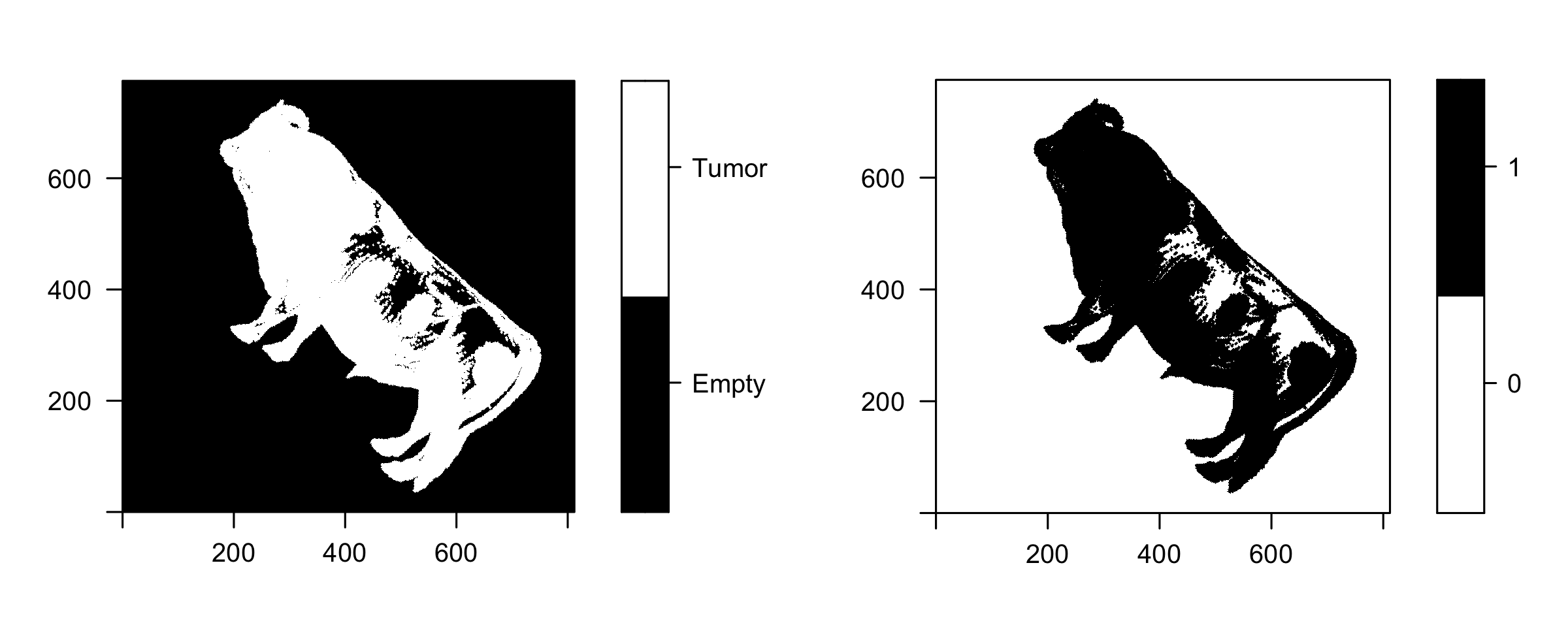
SAFARI (Shape Analysis For AI-Segmented Images) provides functionality for image processing and shape analysis. In the context of reconstructed medical images generated by deep learning-based methods and produced from different modalities such as X-ray, Computational Tomography (CT), Magnetic Resonance Imaging (MRI), and pathology imaging, SAFARI offers tools to segment regions of interest and extract quantitative shape descriptors for applications in signal processing, statistical analysis and modeling, and machine learning.
View the websitePathology Analysis Platform
We developed an innovative online framework (I-Viewer) that leverages AI to enhance pathology diagnosis with real-time assistance and collaboration for pathologists, trainees, and researchers.
View the websiteMicrobiome Community Detector
Microbiome Community Detector (MiCoDe) is a web tool that can be used to investigate the community structure of co-occurrence networks estimated from microbiome data. MiCoDe fits either a Bayesian nonparametric weighted stochastic block model (WSBM) or a Bayesian nonparametric weighted stochastic infinite block model (WSIBM) to a fully connected network represented by a graph with weighted edges.
View the websiteOther Tools
We provide and consistently support online tools, prediction models and software packages. You can download them from the website link below.
View the websiteLatest news
May 2024
ChatGPT can extract data from clinical notes
DALLAS – May 08, 2024 – ChatGPT, the artificial intelligence (AI) chatbot designed to assist with language-based tasks, can effectively extract data for research purposes from physicians’ clinical notes, UT Southwestern Medical Center researchers report in a new study. Their findings, published in NPJ Digital Medicine, could significantly accelerate clinical research and lead to new innovations in computerized clinical decision-making aids.
Collaboration

iSEE-Cell Platform
- Online Tools Development
- Algorithms Sharing
- Comments Collection

Computing Server
- Scripts Upload
- Online Analysis Running
- Slurm Job Management

Data Sever
- User File Upload
- Account-based Data
- Public Built-in Data Access
Selected Publications
-
June 2024
iIMPACT: integrating image and molecular profiles for spatial transcriptomics analysis
Current clustering analysis of spatial transcriptomics data primarily relies on molecular information and fails to fully exploit the morphological features present in histology images, leading to compromised accuracy and interpretability. To overcome these limitations, we have developed a multi-stage statistical method called iIMPACT. It identifies and defines histology-based spatial domains based on AI-reconstructed histology images and spatial context of gene expression measurements, and detects domain-specific differentially expressed genes.
-
May 2024
A critical assessment of using ChatGPT for extracting structured data from clinical notes
This study aims to evaluate ChatGPT’s capacity to extract information from free-text medical notes efficiently and comprehensively. We developed a large language model (LLM)-based workflow, utilizing systems engineering methodology and spiral “prompt engineering” process, leveraging OpenAI’s API for batch querying ChatGPT.
-
December 2023
Deep learning of cell spatial organizations identifies clinically relevant insights in tissue images
In this study, we introduce Ceograph, a cell spatial organization-based graph convolutional network designed to analyze cell spatial organization (for example,. the cell spatial distribution, morphology, proximity, and interactions) derived from pathology images. Ceograph identifies key cell spatial organization features by accurately predicting their influence on patient clinical outcomes.
-
August 2023
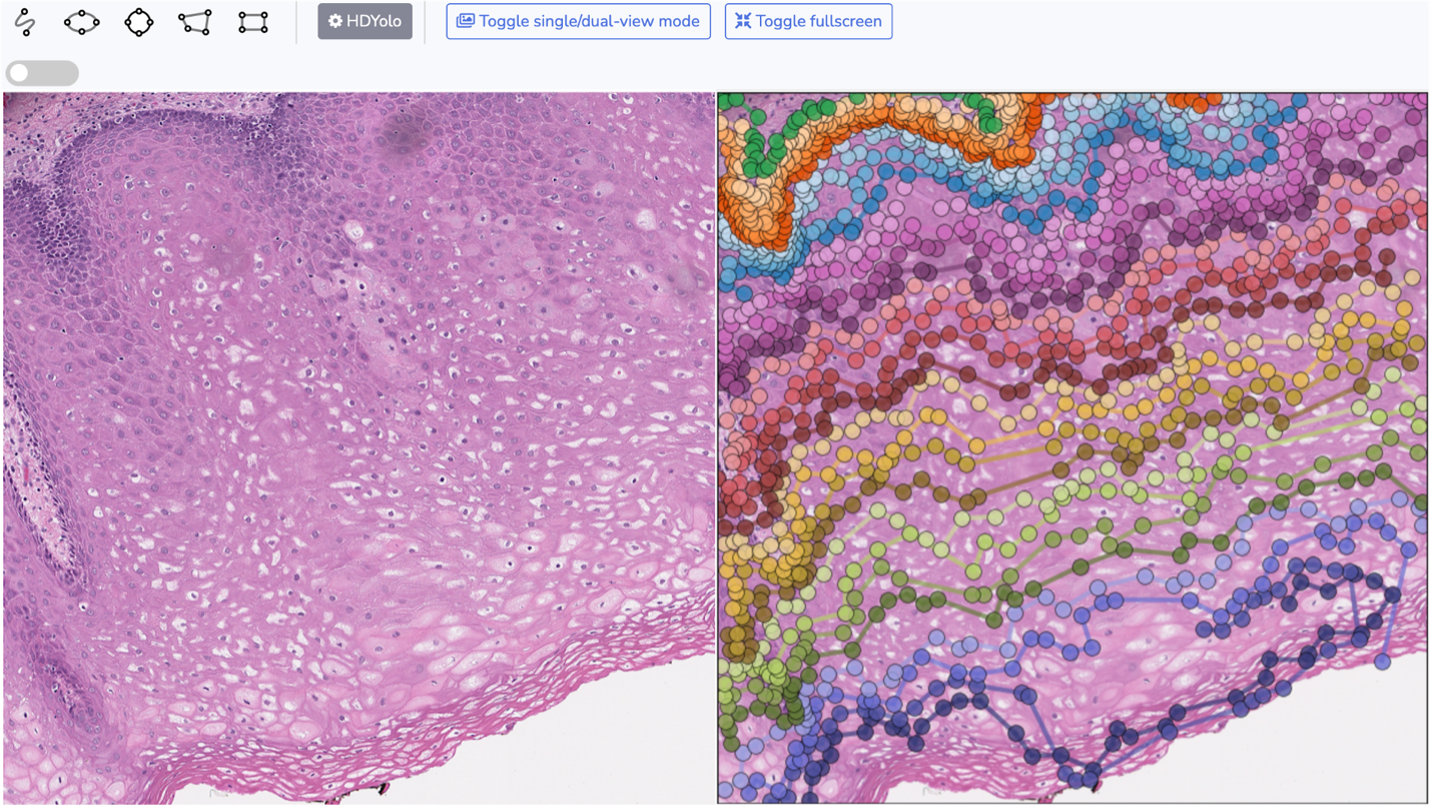
A Deep Learning Approach for Histology-Based Nucleus Segmentation and Tumor Microenvironment Characterization
This study presents Histology-based Detection using Yolo (HD-Yolo), a new method that significantly accelerates nucleus segmentation and TME quantification. We demonstrate that HD-Yolo outperforms existing WSI analysis methods in nucleus detection, classification accuracy, and computation time.
-
June 2022
Deep Learning of Rhabdomyosarcoma Pathology Images for Classification and Survival Outcome Prediction
A convolutional neural network for RMS histology subtype classification was developed using digitized pathology images from 80 patients collected at time of diagnosis. A subsequent embryonal rhabdomyosarcoma (eRMS) prognostic model was also developed in a cohort of 60 eRMS patients.
-
January 2021
A deep learning-based model for screening and staging pneumoconiosis
This study aims to develop an artificial intelligence (AI)-based model to assist radiologists in pneumoconiosis screening and staging using chest radiographs. The model, based on chest radiographs, was developed using a training cohort and validated using an independent test cohort.
-
August 2020
Spatial molecular profiling: platforms, applications and analysis tools
Molecular profiling data generate deep characterizations of the genetic, transcriptional and proteomic events of cells, while tissue images capture the spatial locations, organizations and interactions of the cells together with their morphology features.
-
August 2020
Molecular differences across invasive lung adenocarcinoma morphological subgroups
Lung adenocarcinomas (ADCs) show heterogeneous morphological patterns that are classified into five subgroups: lepidic predominant, papillary predominant, acinar predominant, micropapillary predominant and solid predominant.
-
May 2020
Computational staining of pathology images to study the tumor microenvironment in lung cancer.
These findings present a deep learning-based analysis tool to study the TME in pathology images and demonstrate that the cell spatial organization is predictive of patient survival and is associated with gene expression.
-
December 2019
ConvPath: A software tool for lung adenocarcinoma digital pathological image analysis aided by a convolutional neural network
The spatial distributions of different types of cells could reveal a cancer cell's growth pattern, its relationships with the tumor microenvironment and the immune response of the body, all of which represent key “hallmarks of cancer”. However, the process by which pathologists manually recognize and localize all the cells in pathology slides is extremely labor intensive and error prone.
Preprints
-
arXiv preprint arXiv:2012.04878
Discovering Clinically Meaningful Shape Features for the Analysis of Tumor Pathology Images
EF Morales, C Zhang, G Xiao, C Moon, Q Li
-
arXiv preprint arXiv:2012.12102
Predicting survival outcomes using topological features of tumor pathology images
C Moon, Q Li, G Xiao
-
arXiv preprint arXiv:2012.03326
Bayesian Modeling of Spatial Molecular Profiling Data via Gaussian Process
Q Li, M Zhang, Y Xie, G Xiao
-
arXiv e-prints, arXiv: 2012.04878
Discovering Clinically Meaningful Shape Features for the Analysis of Tumor Pathology Images
E Fernández Morales, C Zhang, G Xiao, C Moon, Q Li
-
arXiv e-prints, arXiv: 2012.04878
Bayesian Landmark-based Shape Analysis of Tumor Pathology Images
arXiv preprint arXiv:2012.01149
Contact Us
Address
Danciger Research Building, 5323 Harry Hines Blvd. Ste. H9.124, Dallas, TX 75390-8821
Phone number
214-648-4110
Guanghua.Xiao@UTSouthwestern.edu
Lab Website
Guanghua Xiao Lab