PROF. GUANGHUA (ANDY) XIAO
Guanghua Xiao is a Professor at the O'Donnell School of Public Health and has secondary appointments in the Departments of Bioinformatics and Biomedical Engineering at UT Southwestern Medical Center. He holds the Mary Dees McDermott Hicks Chair in Medical Science and leads the Health Data Sciences Ph.D. program at the school.
With a focus on machine learning, spatial statistics, and high-dimensional data analysis, Dr. Xiao and his team have developed user-friendly software and tools for genomic and imaging data analysis. His current research includes AI methods for pathology imaging analysis and bioinformatics tools for spatial molecular profiling.
Dr. Xiao is the PI or MPI on several research grants, funded by the National Cancer Institute (NCI), the National Institute of General Medical Sciences (NIGMS), the National Institute of Dental and Craniofacial Research (NIDCR), and the Cancer Prevention and Research Institute of Texas (CPRIT).
Read MoreResearch Summary
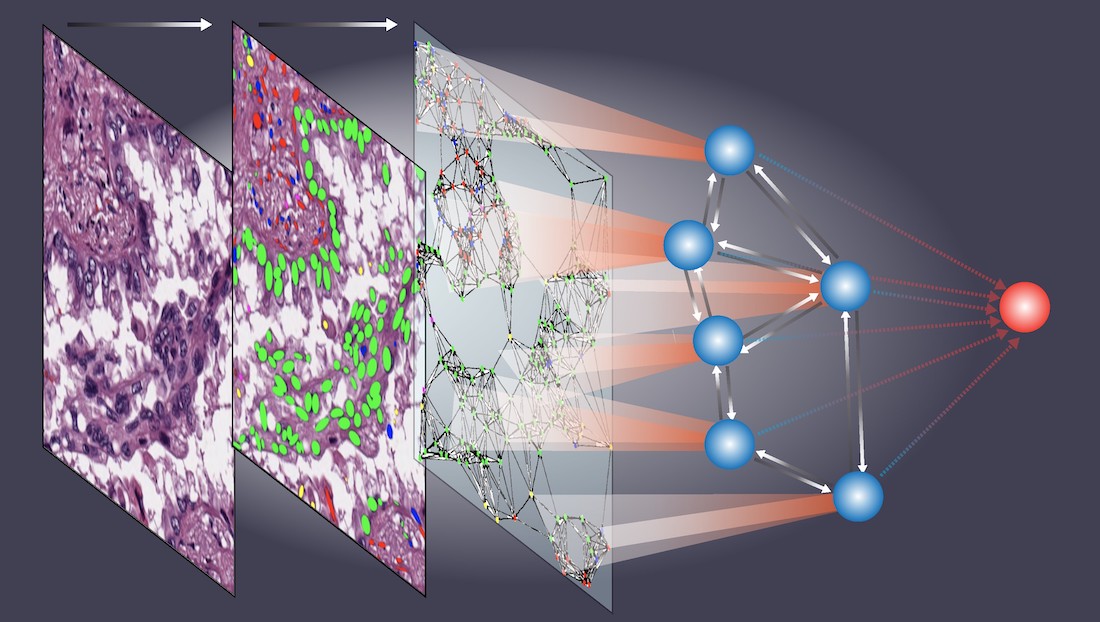
I am interested in developing computational models and algorithms for big data to predict patients' outcomes, which can help clinicians to tailor treatment plans for individual patients. My research lab has developed statistical methodologies for Bayesian analysis, new bioinformatics tools, and gene expression signatures to predict patient prognosis and response to chemotherapy. We are actively developing new bioinformatics tools and computational algorithms for big data, such as imaging and genomic data. Currently, my research is mainly focused on tissue image analysis. Our team was among the first teams to develop and validate computational models using histopathology images collected from routine clinical procedures to refine lung cancer prognosis (Luo et al, 2017, Luo et al, 2019). We have developed deep learning-based models to detect tumor regions, micro-blood-vessels and predict patient outcomes (Wang et al, 2018; Yi et al, 2018; Huang et al, 2017). Recently, we developed algorithms to detect and classify different types of cells from tissue images (Wang et al, 2019; Wang et al, 2020) and developed a set of spatial models (Li et al, 2019, Li et al, 2020) to investigate cell spatial organizations and their implication in disease. Aside from my research in developing statistical methodology, I am also devoted to applying state-of-the-art statistical methods in biomedical research.
Research Interests
Tissue imaging data analysis
Machine learning and deep learning
Developing computational algorithms and bioinformatics tools for complex biomedical data
Developing spatial models for biological data
Biomedical imaging analysis
Selected Publications
Based on an analysis of two large databases on breast cancer, reduced activity of an autophagy gene, beclin 1, was related to both a higher incidence of triple-negative breast cancer and a poorer prognosis for breast cancer patients.
MORE PUBLICATIONSGenomic regression analysis of coordinated expression.
Nature Communications 8, Article number: 2187 (2017)
doi:10.1038/s41467-017-02181-0
Cai L, Li Q, Du Y, Yun J, Xie Y, DeBerardinis RJ, Xiao G.
Using functional signature ontology (FUSION) to identify mechanisms of action for natural products.
Science Signal, Vol. 6, Issue 297, pp. ra90
Potts MB, Kim HS, Fisher KW, Hu Y, Carrasco YP, Bulut GB, Ou
YH, Herrera-Herrera ML, Cubillos F, Mendiratta S, Xiao G,
Hofree M, Ideker T,
Xie Y, Huang LJ, Lewis RE, MacMillan JB, White MA.
Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation.
Science,
Vol. 338, Issue 6109, pp. 956-959
DOI: 10.1126/science.1225967
Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat
G, White M, Reichelt J, Levine B.
News
Try Out Our Software
We have developed online analysis tools which allow users to explore and analyze lung cancer- and germ cell tumor-related gene expression data. PIPECLIP Galaxy is also provided here for biologists to identify the most likely cross-linking sites.