Introduction
The PCDC is funded by CPRIT. Users should acknowledge CPRIT RP180805 on publications supported by this core.
Services :
(1) Provide data sharing, management, and analytics services through a pediatric cancer data commons.
(2) Provide customized data-related service for individual project.
Pediatric Cancer Data Core provide services for
Data Findable, Accessible,
Interoperable, Reusable (FAIR) digital compliance model
Pediatric Cancer Data Commons
We will follow a FAIR digital compliance model (Findable, Accessible, Interoperable, Reusable) to construct the pediatric cancer data commons with high quality and comprehensive data, and provide hardware/software support for users to use this data commons.
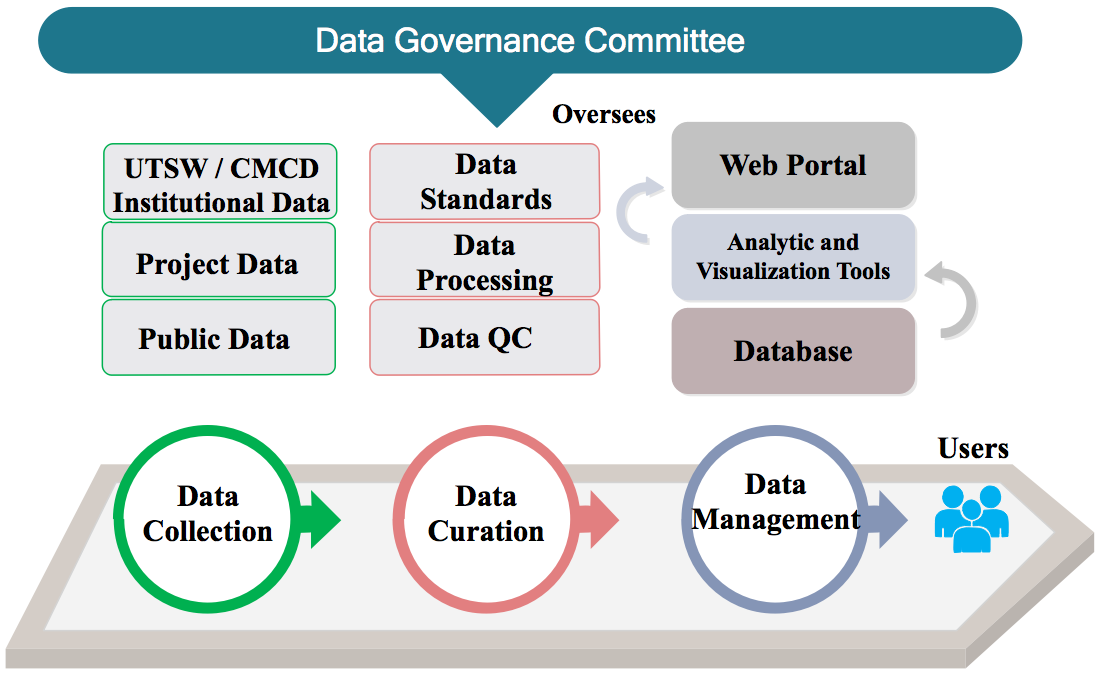
(A) Data Collection
Three types of data sources for the data commons:
(1) UTSW/CMCD Institution-wide data
The PCDC will work with different groups at UTSW/CMCD to collect EHR, clinical trial data, and
sample inventory information for children with cancers. The PCDC will create a virtual sample bank to track
samples and link to clinical and genomic data. Since tumor tissue slides at CMCD’s tissue bank have not been
systematically digitalized, the PCDC will identify and scan these tissue slides, and store the data in our database.
Clinical sequencing data for patient diagnosis will be contributed by UTSW’s clinical NGS lab or external CLIA lab.
All UTSW/CMCD researchers can access these data for free. Data access from external users will be determined
on an individual case level.
(2) Project data
Investigators can contribute their pediatric cancer research data to
the data commons. For example, Dr. Philip Lupo from Baylor College of Medicine plans to use this data
commons to store and share the whole-exome sequencing and phenotype data generated from a CPRIT-funded
molecular epidemiology study (see use cases). Another example is the germ cell tumor clinical trial data from
the MaGIC internal consortium. The access to the project data will follow the
requirements and policies of individual projects.
(3) Public data
The PCDC will identify, collect and curate
pediatric cancer data from the public domain, including
Genomic Data Commons, cBioportal, published papers and
other data repositories. The access to this data will be free to
all users and will follow the data use policy.
(B) Data Curation
To ensure data quality, the PCDC will develop a comprehensive data curation, processing and quality control (QC) procedure for all data collected. We will work closely with clinical experts, users and data standards experts to design a data dictionary, code book and data elements based on the needs from practical use and national data standards. One successful case is that we have been collaborating with MaGIC and CDISC to develop a data dictionary for pediatric germ cell tumors and used it to collect clinical and genomic data from international clinical trials. The PCDC will set a standard operating procedure (SOP) and code pipelines for the data curation and QC. The SOP and codes will be well documented using Github and other version control tools and open to all the users.
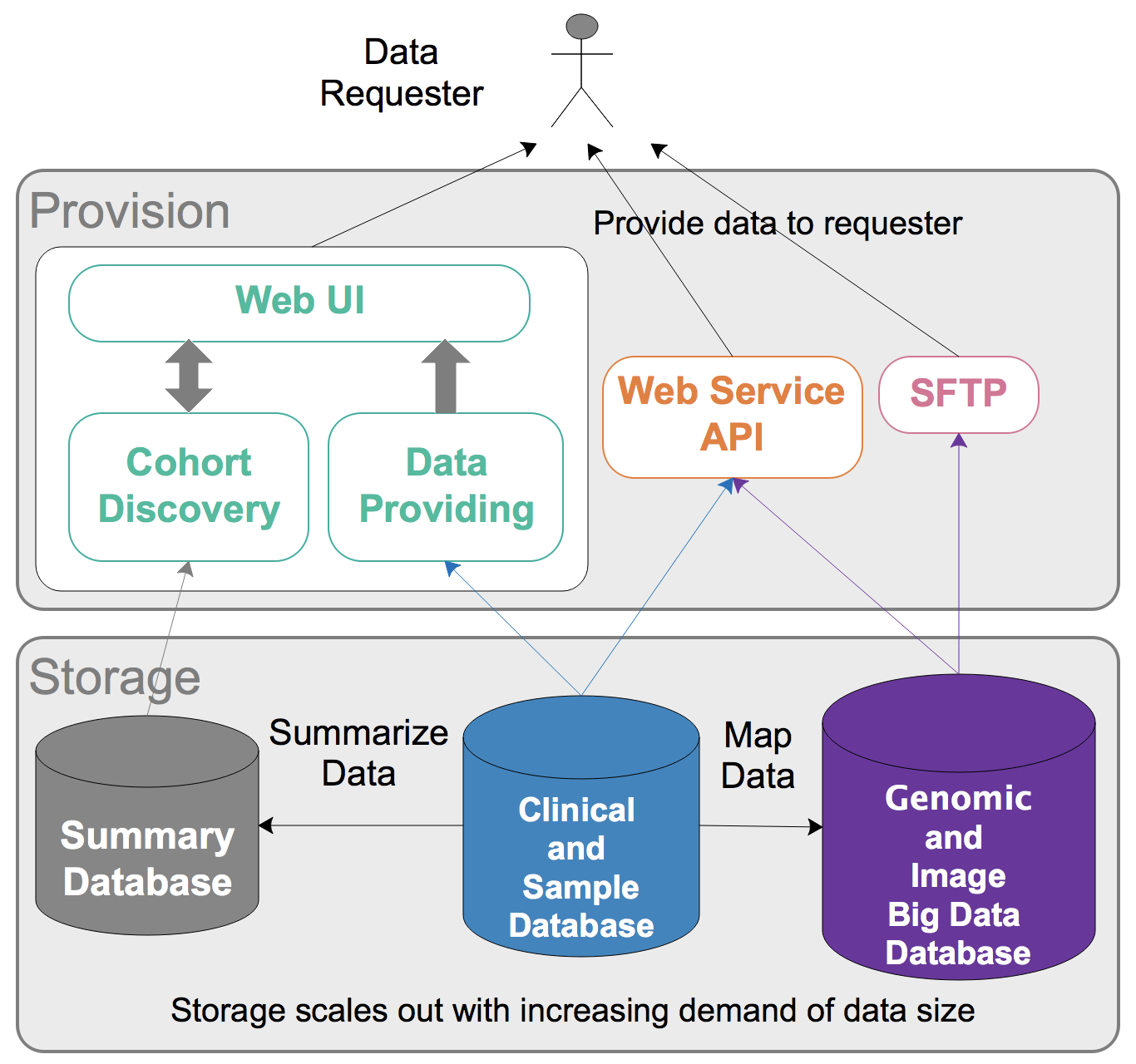
(C) Data Management
Developing a secure, robust and scalable data storage and organization infrastructure is the key for a data commons. We will develop cloud-based data storage and analysis toolsets as well as web portals with user-friendly interface. Additionally, different tools will be developed to facilitate user access and data analysis. Summary statistics of data availability from UTSW, collaborative projects and pubic data will be presented in our cohort discovery portal, and all users can easily access it. Access to individual-level data will follow the data governance plan and require appropriate approvals.
(D) Hardware and Software Support
The data common users will receive access to the BioHPC – a fully integrated, modular, and scalable computing facility at UTSW. The users will be supported by the PCDC to access 1,500 TB storage space and high performance computing environment to meet the demand of their research. We will subscribe to a variety of software and support tools based on user demand and past use statistics. The toolsets will include biospecimen management (Open Specimen), data importing/exporting (RedCap), cloud computing (OpenStack), text mining and NLP (MedEx, CARD and CLAMP), and many other tools. Users could get access to these tools for free or with a substantial discount and full user support from PCDC.
(E) Project Support
PCDC will implement two programs to provide customized support to individual pediatric cancer-related research across campus and Texas:
(1) Data science help desk:
Core staff members will be available for consultations, including assistance with
hardware, software, study design and data analysis. CPRIT funds will be allocated to offer this service for free.
(2) Collaboration:
For larger projects, core staff can be engaged through contributions of appropriate FTEs. The goal
of this service is to provide the pediatric cancer-research community with data science personnel on demand;
i.e., a lab will have access to highly qualified data scientists for a defined period of time, without the challenges
of recruiting and retaining such personnel. CPRIT funds will be used for the initial recruitment of qualified
personnel and for bridge funding in the early phases of the program where not all FTEs are fully covered by
grants. With CPRIT funds we anticipate fostering strong interactions between the PCDC and the user community
that guarantee self-sustained continuation of the collaboration program after sunset of the grant award.
| Program | Service |
|---|---|
| Data Science Help Desk |
|
| Collaboration |
|