Clinical Databases
COVID19 Immune Viewer
The outbreak of the 2019 Novel Coronavirus (SARS-CoV-2) rapidly spread from Wuhan, China to multiple countries, causing staggering number of infections and deaths. A systematic profiling of the immune vulnerability landscape of SARS-CoV-2, which can bring critical insights into the immune clearance mechanism, peptide vaccine development, and antiviral antibody development, is lacking. In this study, we investigated the potential of the SARS-CoV-2 viral proteins to induce class I and II MHC presentation and to form linear antibody epitopes. We created an online database to broadly share the predictions as a resource for the research community. Using this resource, we showed that genetic variations in SARS-CoV-2, though fewer for the moment, already follow the pattern of mutations in related coronaviruses, and could alter the immune vulnerability landscape of this virus. Importantly, we discovered evidence that SARS-CoV-2 used mutations to evade attack from the human immune system. Overall, we present an immunological resource for SARS-CoV-2 that could significantly promote both therapeutic development and mechanistic research.
Link: https://qbrc.swmed.edu/projects/2019ncov_immuneviewer/
GCTs
Germ Cell Tumors (GCTs) are malignant cancers that arise from embryonic precursors known as Primordial Germ Cells. GCTs occur in neonates, children, adolescents and young adults and can occur in the testis, the ovary or extragonadal sites. Because GCTs arise from pluripotent cells, the tumors can exhibit a wide range of different histologies. Current cisplatin-based combination therapies cure most patients, however at the cost of significant toxicity to normal tissues. While GWAS studies and genomic analysis of human GCTs have uncovered some somatic mutations and loci that might confer tumor susceptibility, little is still known about the exact mechanisms that drive tumor development.

Leukemia Explorer
Preliminary Demonstration for Acute Lymphoblastic Leukemia. (Data Source: Target)
Neuroblastoma Explorer
Preliminary Demonstration for Neuroblastoma. (Data Source: Target)
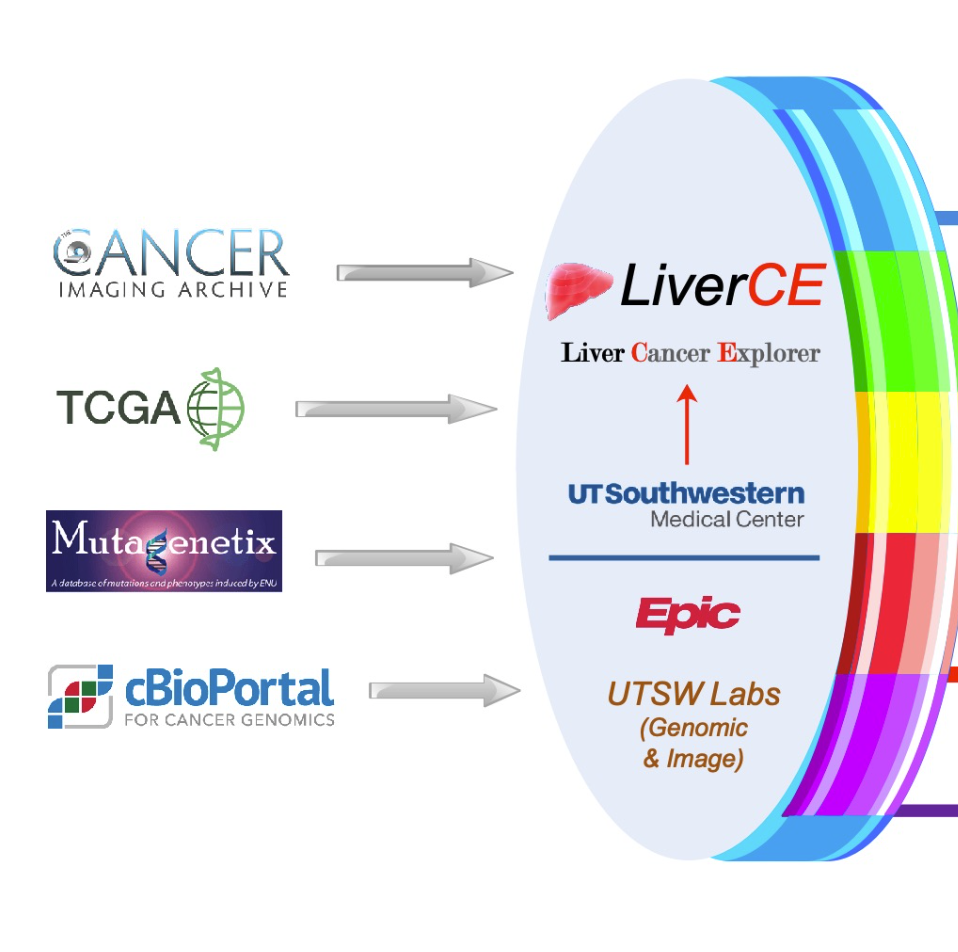
Liver Cancer Explorer
The University of Texas Southwestern (UTSW) Liver Cancer Explorer (LiverCE) leverages a rich array of in-house basic science discoveries for their seamless translation to the care of patients with liver cancer, which is the fastest rising cause of cancer mortality in the U.S. and the most prevalent in Texas. LiverCE has assembled a uniquely diverse multidisciplinary team of basic, clinical, and translational scientists to tackle this major health problem, which disproportionately affects the racially/ethnically and socioeconomically diverse patient population in our catchment area in North Texas.
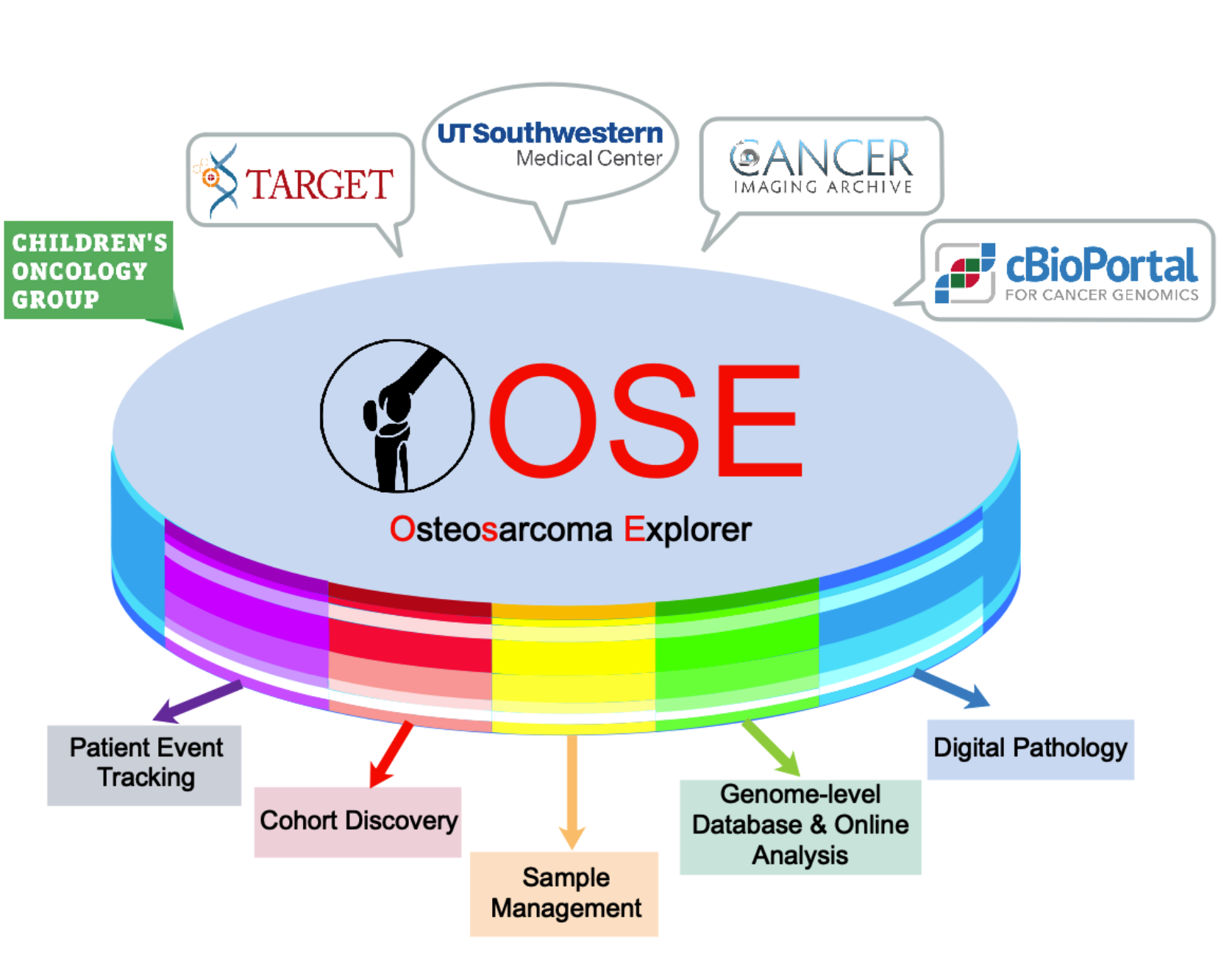
Osteosarcoma Explorer
Osteosarcoma Explorer (OSE) incorporates a rich array of scientific discoveries for their translation to the care of osteosarcoma patients. The OSE project has assembled a multidisciplinary team of clinical, basic, and data scientists to tackle this rare yet severe disease.

Pediatric PDX Explorer
Patient Derived Xenograft (PDX) Explorer incorporates a rich array of scientific discoveries for their translation to the care of Pediatric patients. The Pediatric PDX Explorer project has assembled a multidisciplinary team of clinical, basic, and data scientists to tackle rare yet debilitating diseases.
Lung Cancer Explorer
We constructed a lung cancer-specific database housing expression data and clinical data from over 6700 patients in 56 studies. Expression data from 23 genome-wide platforms were carefully processed and quality controlled, whereas clinical data were standardized and rigorously curated. Empowered by this lung cancer database, we created an open access web resource—the Lung Cancer Explorer (LCE), which enables researchers and clinicians to explore these data and perform analyses. Users can perform meta-analyses on LCE to gain a quick overview of the results on tumor vs non-malignant tissue (normal) differential gene expression and expression-survival association. Individual dataset-based survival analysis, comparative analysis, and correlation analysis are also provided with flexible options to allow for customized analyses from the user.

Microbiome Gym
Microbiome Gym hosts simulated datasets with documented assumptions, codes and real datasets with references. It provides a wide range of statistical tests and their benchmarking. We encourage peers to share their method development results and benchmark openly.

cBioPortal
The cBioPortal for Cancer Genomics provides visualization, analysis and download of large-scale cancer genomics data sets.
Link: https://qbrc2.swmed.edu/
