Data Visualization
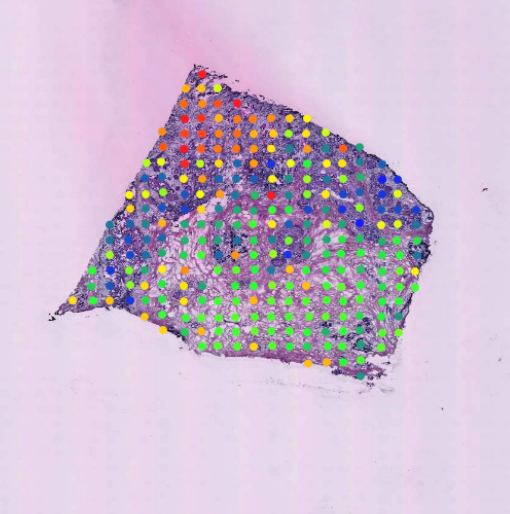
Spatial Transcriptomics Arena
The Spatial Transcriptomics Arena(STAR) is an open platform for methodology research. It provides resources including curated datasets from simulation and real studies (Dataset tab), accessible analysis methods (Method tab), and reproducible analysis results (Analysis tab). It also welcomes new datasets, methods, or analysis results contributed by the broad research community.

ScopeViewer
A convenient web-based Image Viewer for high-resolution wholeslides. Put data anywhere safe and view it on our website.

I-Viewer
I-Viewer is a role-based management system with Authentication and Authorization for image viewing and annotation.
Link: https://digital-pathology.swmed.edu/i-viewer/
Note: Only access in UTSW campus or through VPN from off-campus

Pediatric PDX Explorer
Patient Derived Xenograft (PDX) Explorer incorporates a rich array of scientific discoveries for their translation to the care of Pediatric patients. The Pediatric PDX Explorer project has assembled a multidisciplinary team of clinical, basic, and data scientists to tackle rare yet debilitating diseases.
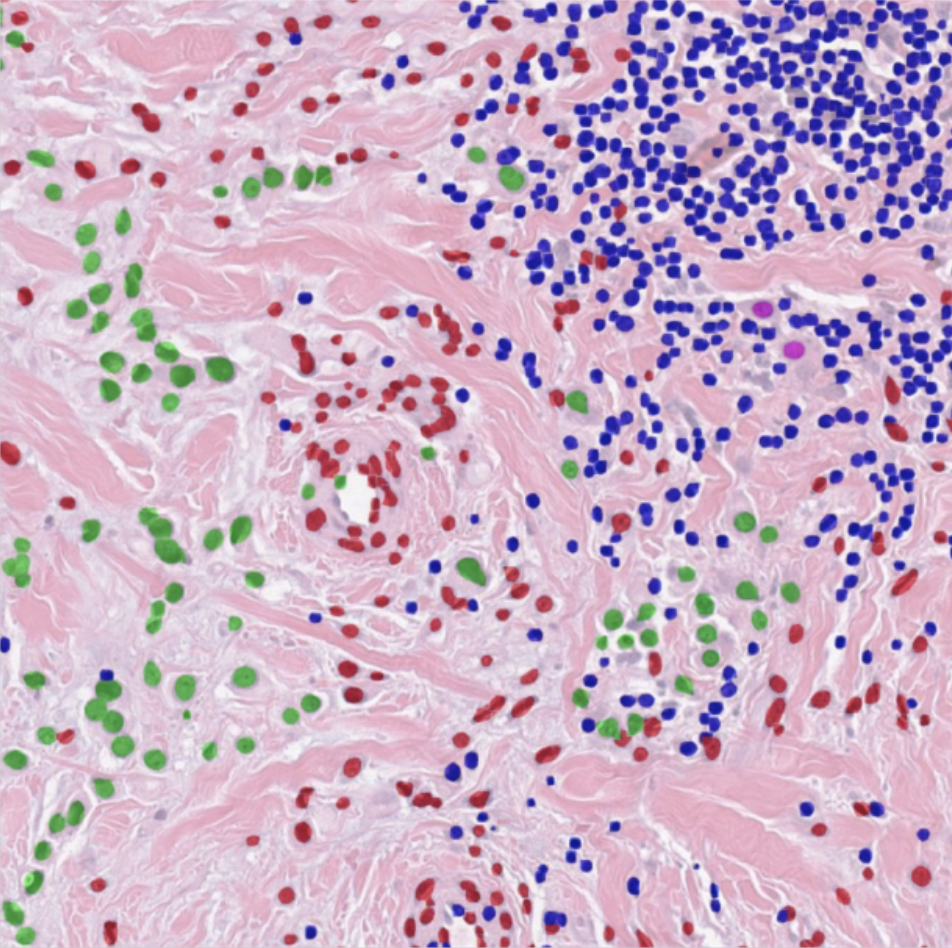
HD-staining v2
The HD-Staining-v2 system utilize the latest one-stage detection algorithms YOLO to dramatically enhance the nuclei detection and segmentation speed. The new system is 15 times faster than HD-Staining-v1 with improved accuracy and precision. Whole slide imaging analysis reveals the blazing fast speed of the v2 system that can potentially reduce the workloads from weeks to hours.
Rhabdomyosarcoma Explorer
Rhabdomyosarcoma (RMS) Explorer incorporates a rich array of scientific discoveries for their translation to the care of pediatric rhabdomyosarcoma patients. The RMSE project has assembled a multidisciplinary team of clinical, basic, and data scientists to tackle rare yet debilitating diseases.
Link: http://lce-test.biohpc.swmed.edu/rms/
Note: Only access in UTSW campus or through VPN from off-campus
Lung Cancer Explorer
We constructed a lung cancer-specific database housing expression data and clinical data from over 6700 patients in 56 studies. Expression data from 23 genome-wide platforms were carefully processed and quality controlled, whereas clinical data were standardized and rigorously curated. Empowered by this lung cancer database, we created an open access web resource—the Lung Cancer Explorer (LCE), which enables researchers and clinicians to explore these data and perform analyses. Users can perform meta-analyses on LCE to gain a quick overview of the results on tumor vs non-malignant tissue (normal) differential gene expression and expression-survival association. Individual dataset-based survival analysis, comparative analysis, and correlation analysis are also provided with flexible options to allow for customized analyses from the user.
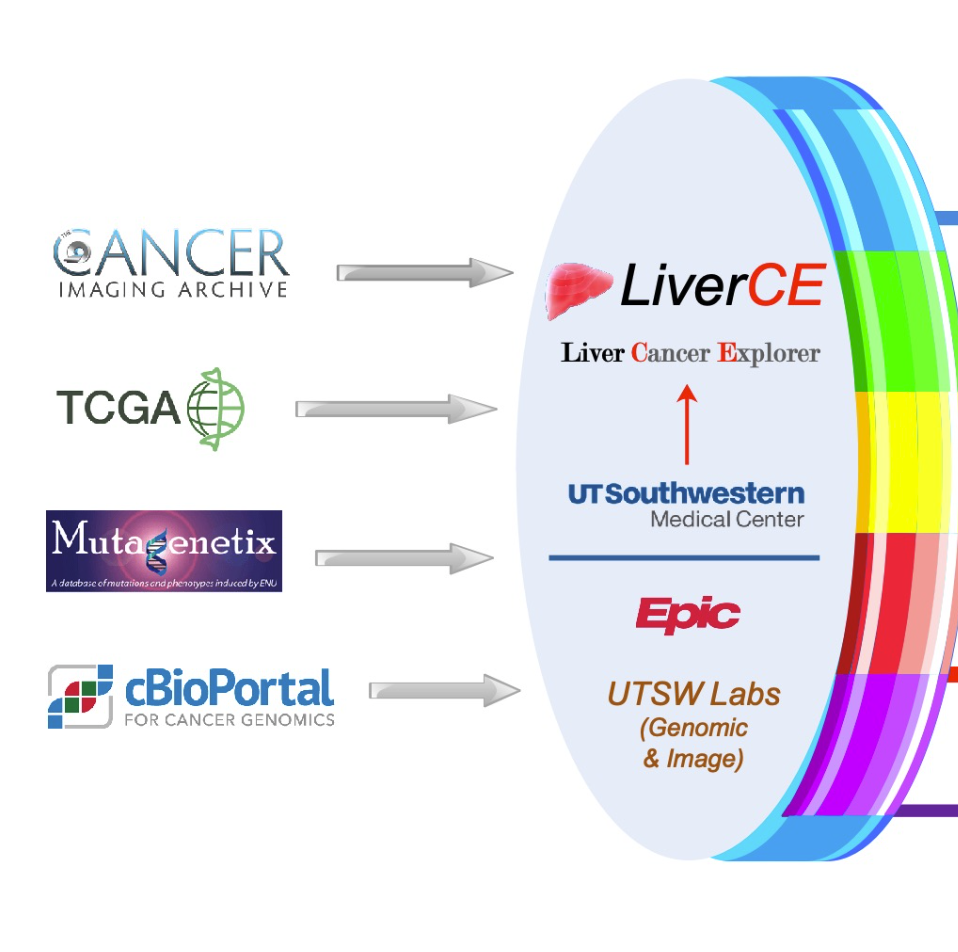
Liver Cancer Explorer
The University of Texas Southwestern (UTSW) Liver Cancer Explorer (LiverCE) leverages a rich array of in-house basic science discoveries for their seamless translation to the care of patients with liver cancer, which is the fastest rising cause of cancer mortality in the U.S. and the most prevalent in Texas. LiverCE has assembled a uniquely diverse multidisciplinary team of basic, clinical, and translational scientists to tackle this major health problem, which disproportionately affects the racially/ethnically and socioeconomically diverse patient population in our catchment area in North Texas.
SARS-CoV-2
We investigated the potential of the SARS-CoV-2 viral proteins to induce class I and II MHC presentation and to form linear antibody epitopes. This online database is established to broadly share the predictions as a resource for the research community.
Link: http://lce-test.biohpc.swmed.edu/ncov/
Note: Only access in UTSW campus or through VPN from off-campus
